Mechanics and Signalling in Phagocytosis
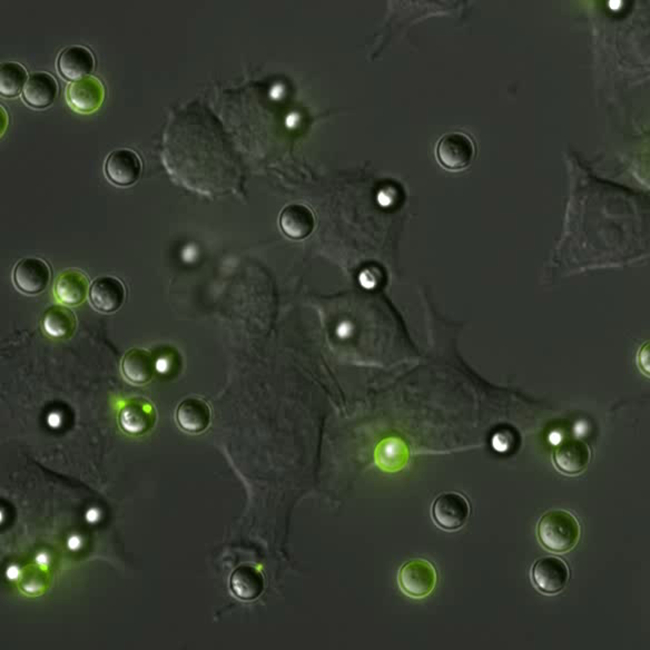 Phagocytosis is the process by which cells engulf foreign bodies. The process is ubiquitous throughout nature. The behavior can be observed in unicellular organisms like amoeba, which phagocytose their prey. Similarly, for vertebrates phagocytosis is the hallmark method white blood cells (macrophages and neutrophils) use to eliminate pathogens and debris from the body.
Phagocytosis is the process by which cells engulf foreign bodies. The process is ubiquitous throughout nature. The behavior can be observed in unicellular organisms like amoeba, which phagocytose their prey. Similarly, for vertebrates phagocytosis is the hallmark method white blood cells (macrophages and neutrophils) use to eliminate pathogens and debris from the body.
Understanding the mechanisms that drive phagocytosis in mammalian cell types can provide information assisting in the treatment of pathogens like listeria which target cells through the phagocytic pathway and other immune response related complications such as inflammation, heart disease and cancer proliferation.

Fc-Receptor Mediated Phagocytosis:
The primary method by which macrophages and neutrophils identify targets is through the use of specialized receptors in their membrane. In the case of Fc-mediated phagocytosis, the process consists of four general steps (pictured right):- Opsonization of target with IgG, forming an immune complex (IC)
- Binding of the IC to Fc-Receptors on the cell’s surface
- Clustering of the Fc-Receptors around the IC, increasing the signal
- Initiation of phagocytosis, in which the cell engulfs the IC
Phagocytosis Related Bio-machinery
There are a number of distinct physical processes that drive phagocytosis. These include both “passive” processes such as changes in target-membrane wettability due to receptor binding, and “active” processes like the formation of cellular protrusions driven by actin polymerization.Receptor Binding & Wettability:

 Simplifying the target particle, cell and extracellular fluid into a 3-phase system, the Young equation specifies the contact angle as a function of interfacial tensions.
Simplifying the target particle, cell and extracellular fluid into a 3-phase system, the Young equation specifies the contact angle as a function of interfacial tensions.In this limited model, higher cell-target affinities will drive the bead into the body.
Membrane Expansion:
 For a cell to engulf a target it must increase its surface area. The increase in surface area is believed to come from exocytosis of vesicles within the cell body, and unfolding of micro-wrinkles in the membrane. The work of Evans et al. and Heinrich et al. have demonstrated a non-linear increase in membrane cortical tension, which suggests that the phagocytic pathway at some point triggers the release of tightly bound micro-wrinkles.[1,2]
For a cell to engulf a target it must increase its surface area. The increase in surface area is believed to come from exocytosis of vesicles within the cell body, and unfolding of micro-wrinkles in the membrane. The work of Evans et al. and Heinrich et al. have demonstrated a non-linear increase in membrane cortical tension, which suggests that the phagocytic pathway at some point triggers the release of tightly bound micro-wrinkles.[1,2]
Actin Polymerization:
 The requirement of actin polymerization is documented throughout the literature.[3] Blocking actin polymerization with the drug cytochalasin results in decreased phagocytic uptake[3,4]. However, the structure of the polymerized actin and the role it plays are still an active area of research. There are models that propose that actin meshwork forms a scaffolding that prevents the cell membrane from “slipping backwards” as it wicks around a target[5]. There are other models that suggest it assists in the closure of the phagosome[6].
The requirement of actin polymerization is documented throughout the literature.[3] Blocking actin polymerization with the drug cytochalasin results in decreased phagocytic uptake[3,4]. However, the structure of the polymerized actin and the role it plays are still an active area of research. There are models that propose that actin meshwork forms a scaffolding that prevents the cell membrane from “slipping backwards” as it wicks around a target[5]. There are other models that suggest it assists in the closure of the phagosome[6].
Myosin Activity:
 There are reports of phagocytosis being affected by myosin inhibitors. It is unclear what the role of these molecular motors are; however, it have been shown that myosin classes I, II, V and IX are present during phagocytosis in mouse derived macrophages. It has also been shown that myosin IC localized to the leading edge of phagosomes as they close around the target particle[6].
There are reports of phagocytosis being affected by myosin inhibitors. It is unclear what the role of these molecular motors are; however, it have been shown that myosin classes I, II, V and IX are present during phagocytosis in mouse derived macrophages. It has also been shown that myosin IC localized to the leading edge of phagosomes as they close around the target particle[6].
This material is based upon work supported by the National Science Foundation under Grant No. 0848797.
Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
References:
[1] Heinrich, et al. J. Cell. Sci. 118, 1789-1797 (2005)
[2] Evans, et al. Biophys. J. 56, 151-160 (1989)
[3] Hirsch et al. Exp. Cell Res. 73, 383-393 (1972)
[4] Gridstein et al. J. Biol. Chem. 278, 3331-3338 (2003)
[5] Endres et al. BMC Sys. Bio. 4:149 (2010)
[6] Swanson et al. J. Cell Sci. 112, 307-316 (1999)
[7] Rottner et al. EMBO J. 27, 982-992 (2008)